About¶
About TIMSCONVERT¶
TIMSCONVERT is a workflow designed for mass spectrometrists that allows for the conversion of raw data from Bruker timsTOF series mass spectrometers (i.e. Pro, fleX, SCP, HT, etc.) to open data formats (i.e. mzML and imzML) that:
are compatible with downstream open source data analysis platforms.
incorporate trapped ion mobility spectrometry (TIMS) data into these open formats.
does not require any programming experience.
timsTOF File Formats¶
All Bruker data is housed in a directory with the .d extension. Within the directory, the acquisition software and type
of experiment conducted (i.e. acquisition mode) determines the data format. LC-MS(/MS) experiments acquired using
Bruker otofControl use the BAF file format, which is widely supported by both proprietary (i.e. Bruker DataAnalysis,
CompassXport) and open-source (i.e. Proteowizard MSConvert) data conversion/analysis software. The raw binary data is
housed in the analysis.baf file, while the metadata and recalibration information are stored in SQLite databases
titled analysis.sqlite and calibration.sqlite, respectively.
LC-MS(/MS) and MALDI-MS(/MS) dried droplet and mass spectrometry imaging experiments acquired using Bruker timsControl
use the TSF file format. Compared to BAF files, TSF files have the advantage of store data in a much more efficient
manner, but have more limited support from both proprietary and open-source software. Experiments designed to acquire
TIMS data (LC-TIMS-MS(/MS), MALDI-TIMS-MS(/MS) dried droplet and mass spectrometry imaging) use the TDF file formats,
which are similar to TSF files but also contain TIMS data. Run metadata are stored in the analysis.tsf/
analysis.tdf SQLite databases, raw binary data are stored in the analysis.tsf_bin/analysis.tdf_bin files,
and recalibration data are stored in the calibration.sqlite SQLite database.
Dimensionality of timsTOF Data¶
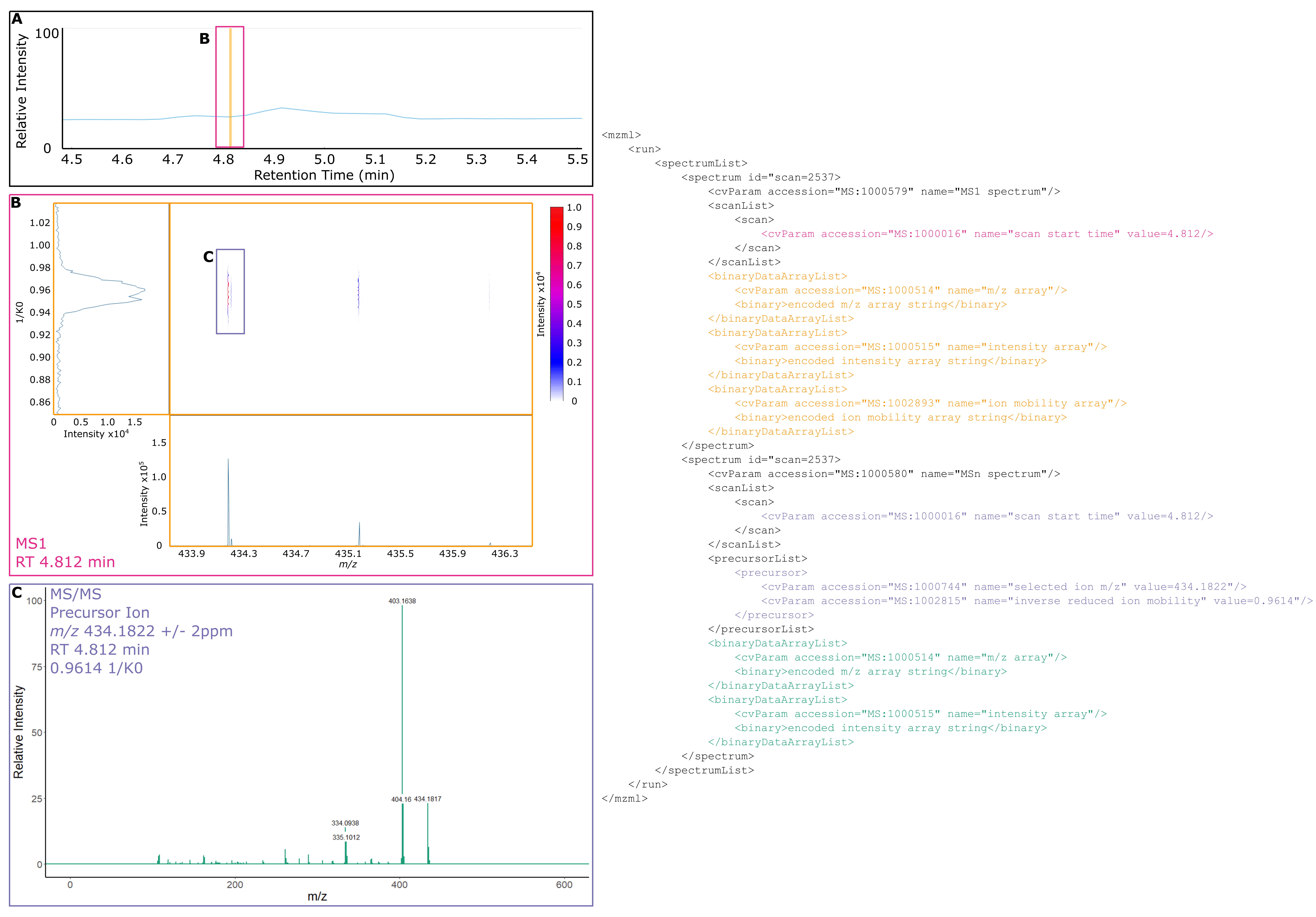
Example of dimensionality of LC-TIMS-MS/MS data and a simplified corresponding mzML schema. Elements in the simplified mzML schema are color coded by the corresponding data in the chromatograms/mobilograms/spectra. At a given retention time in the chromatogram (indicated in pink), an MS1 spectrum may be visualized in the form of a three dimensional plot generated from m/z, intensity, and ion mobility arrays (indicated in orange). Precursor ions of interest found in MS1 spectra (indicated in purple) can be further analyzed by plotting m/z and intensity arrays for MS/MS spectra (indicated in teal). These spectral identifiers (i.e. retention time, precursor m/z, precursor 1/K0) may be used to locate the corresponding MS/MS spectrum from Bruker DataAnalysis or other data visualization software of choice.
Example Acquisition Modes for File Conversion¶
LC-MS(/MS) (BAF or TSF) -> mzML
LC-TIMS-MS(/MS) (TDF) -> mzML
MALDI-MS(/MS) Dried Droplet (TSF) -> mzML
MALDI-MS Imaging Mass Spectrometry (TSF) -> imzML
MALDI-TIMS-MS(/MS) Dried Droplet (TDF) -> mzML
MALDI-TIMS-MS Imaging Mass Spectrometry (TDF) -> imzML
Downstream Data Analysis¶
Examples of downstream data analysis platforms include and are not limited to: * Global Natural Products Social (GNPS) * Cardinal MSI * IDBac * MZmine